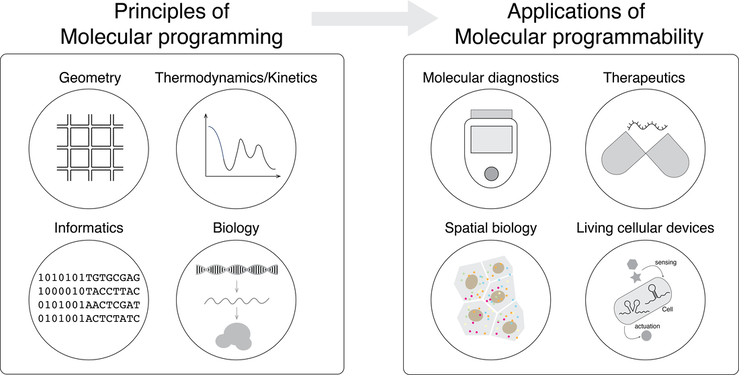
Overview
Biomolecules are the foundations of life science, chemistry, and biomaterials development. The capability to manipulate biomolecules in terms of structures and interaction pathways will open tremendous opportunities in strengthening our understanding of biology, develop novel biomaterials, and revolutionize biotechnologies. The key challenges along this direction are how to understand the molecular interactions in nature, how to design molecular systems de-novo, and how to integrate molecular design with biology and material engineering for next-generation therapeutic, bioanalysis, and biomaterials. To achieve these goals, my research will move along the following areas:
1. Design of Bimolecular structures and dynamics
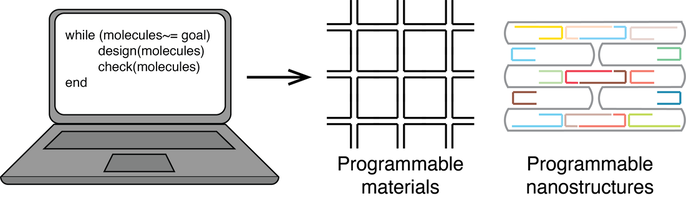
We have thousands of Apps on our computers and cell phones to serve our daily life and work, they are achieved by programming languages in the information world. There are also thousands of molecular apps in our body to maintain numerous cellular activities to keep us healthy.
Here raises the question that can we design those molecule apps as we program Apps in our cell-phone and computer? If we achieved this, we can design molecule robots to cure disease, molecular algorithm for advanced bio-analysis, novel biomaterials, and eventually even to program life. In this direction, I’m interested in designing molecular structures and dynamic systems from scratch to ever-increased complexity and functionality. To achieve this goal, we need to understand how a single molecular folds, how different molecules interact with each other, and how those folding and interaction lead to their functions.
I'll develop both experimental and computational methods to address those challenges. The development principles will lead to the next generation’s molecular machines, novel biomaterials discovery, bioanalysis tool and many more beyond imagination.
Here raises the question that can we design those molecule apps as we program Apps in our cell-phone and computer? If we achieved this, we can design molecule robots to cure disease, molecular algorithm for advanced bio-analysis, novel biomaterials, and eventually even to program life. In this direction, I’m interested in designing molecular structures and dynamic systems from scratch to ever-increased complexity and functionality. To achieve this goal, we need to understand how a single molecular folds, how different molecules interact with each other, and how those folding and interaction lead to their functions.
I'll develop both experimental and computational methods to address those challenges. The development principles will lead to the next generation’s molecular machines, novel biomaterials discovery, bioanalysis tool and many more beyond imagination.
References:
- Fan Hong, John Shreck, Petr Sulc, Understanding DNA interactions in crowded environments with a coarse-grained model, Nucleic Acid Research, 2020,48,10726.
- Fan Hong, Shuoxing Jiang, Xiang Lan, Raghu Pradeep Narayanan, Petr, Sulc, Fei Zhang, Yan Liu, Hao Yan, Layered-Crossover Tiles with Precisely Tunable Angles for 2D and 3D DNA Crystal Engineering, J. Am. Chem. Soc. 2018, 140, 14670-14676.
- Shuoxing Jiang #, Fan Hong#, Huiyu Hu, Hao Yan, Yan Liu, Understanding the Elementary steps in DNA tile-based self-assembly. ACS nano, 2017, 11,9370-9381. Contributed Equally.
- Fan Hong, Fei Zhang, Yan Liu, Hao Yan,DNA origami: scaffolds for creating high order structures. Chemical Review, 2017, 117, 12584-12640
- Fan Hong, Shuoxing Jiang, Tong Wang, Yan Liu, Hao Yan, 3D Framework DNA origami structure with layered crossover motifs. Angew Chem Int Ed, 2016,55, 12832-12835.
- Wei Guo #, Fan Hong#, et al, Fan Xia*. Target-Specific 3D DNA Gatekeepers for Biomimetic Nanopores, Advanced Materials, 2015, 27,2090-2095. Contributed Equally.
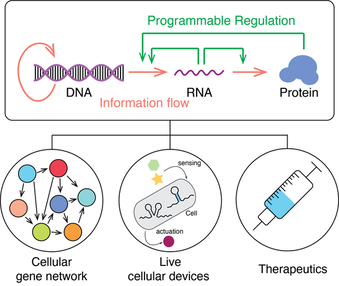
2. Synthetic Biology with designed molecular machines
The central dogma is key regulation of biology. One of the key concepts of synthetic biology is to reprogram the cell through rewiring the central dogma by gene expression regulation. Along this direction, I’ll develop a novel mechanism to hack the centra dogma to program cellular gene expression. Can we design molecular tools to rewire genome, transcriptome, and proteome. The regulation mechanism can be used to develop a complex “bio-algorithm”, develop living cellular devices to record the environmental signal, and novel gene therapeutic methods.
References:
- Fan Hong, Petr Sulc. An emergent understanding of strand displacement in RNA biology, Journal of Structural Biology, 2019, 207, 241-249.
- Fan Hong, Duo Ma, Kaiyue Wu, Lida A. Mina, Rebecca C. Luiten, Yan Liu, Hao Yan*, Alexander A. Green*. Precise and Programmable Detection of Mutations Using Ultraspecific Riboregulators, Cell, 2020, 180, 1018-1032.
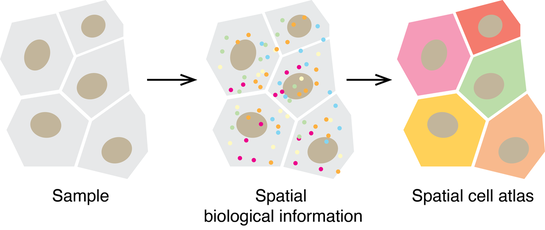
3. Spatial biology with DNA advanced imaging
Cells have their neighbors and will affect each other. Traditional biological analysis will blend all the cells in a tissue sample and the spatial information is missing. To understand the disease and cellular function with insights, we need to use “GPS” to create a map and track each cell within its local context. Along this direction, I’ll develop novel tools to facilitate the reading of spatial information in the biological context with DNA advanced multiplexed imaging.
I'll develop method that significantly expands the multiplexity by using advanced DNA design to visualize biology (e.g., genomics, transcriptomics, and proteomics). The method will transform the way to illuminate biology spatially with unprecedented speed and information to understand biology with precision at the single-cell level and scalable at the tissue scale. The comprehensive spatial biological information will facilitate our understanding of tissue function and human diseases.
I'll develop method that significantly expands the multiplexity by using advanced DNA design to visualize biology (e.g., genomics, transcriptomics, and proteomics). The method will transform the way to illuminate biology spatially with unprecedented speed and information to understand biology with precision at the single-cell level and scalable at the tissue scale. The comprehensive spatial biological information will facilitate our understanding of tissue function and human diseases.
References:
Comming soon
Comming soon
4. Molecular Diagnostics for clinics
Caner and virus infection are become one of the key threats of human and global health. Molecular diagnostics are essential to fight against those bio-threats. Along this direction, I’ll leverage my knowledge of molecular programmability to develop the cutting-edge technologies and apply them to the urgent need of global challenges, such as Zika outbreak, Covid19 pandemic and cancer diseases to achieve advanced diagnostics with faster turn-around time, higher in-depth information, lower economic cost and more reachable in resource limited environment comparing with traditional methods.
References:
- Youngeun Kim, Adam B Yaseen, Jocelyn Y Kishi, Fan Hong, Sinem K Saka, Kuanwei Sheng, Nikhil Gopalkrishnan, Thomas E Schaus, Peng Yin. Single-stranded RPA for rapid sensitive detection of SARS-Cov-2 RNA. MedRxiv.
- Fan Hong, Duo Ma, Kaiyue Wu, Lida A. Mina, Rebecca C. Luiten, Yan Liu, Hao Yan*, Alexander A. Green*. Precise and Programmable Detection of Mutations Using Ultraspecific Riboregulators, Cell, 2020, 180, 1–15
- Xiaowen Ou, Fan Hong, et al, Fan Xia*, A highly sensitive and facile graphene oxide-based nucleic acid probe: Label-free detection of telomerase activity in cancer patient's urine using AIEgens, Biosensors and Bioelectronics, 2016, 89, 417-421.